Day38: Refinement
Today we will continue with the exploration of the pesticides. The data I use comes from Kaggle’s Pesticide Data Program (2015) data set uploaded by the United States Department of Agriculture.
I continued with R and the markdown document for this little project is found here. (You can also find the R Markdown document in the sample directory, but it’s less pretty.)
Factor levels
One heuristics that I generally look at is the number of distinct values a variable take. A low number suggests the variable is grossly divided, possibly a categorical variable, and a high number suggests there are many divisions or the variable is a continuous variable.
sapply(colnames(results), function(var){
results[[var]] %>% unique() %>% length()
}) %>%
{data.frame(var = names(.), n = .)} %>%
arrange(desc(n))
## var n
## 1 sample_pk 10187
## 2 concen 2582
## 3 pestcode 489
## 4 lod 126
## 5 commod 20
## 6 testclass 20
## 7 lab 8
## 8 annotate 5
## 9 determin 5
## 10 confmethod 4
## 11 mean 4
## 12 commtype 3
## 13 quantitate 3
## 14 extract 2
## 15 conunit 1
## 16 confmethod2 1
We find n = 1 for “conunit” and “confmethod2”, which tells you nothing interesting about the data. Such variables can be discarded from further analysis. From the results, I also see that the extraction method (“extract”) takes on 2 values.
Exploring the extraction methods
- 805 = MDA Modified QuEChERS Method
- 818 = NSL Animal Tissue Extraction Method
# Generate separate figures
p1 <-
results %>%
ggplot(aes(x=extract, fill=factor(extract))) +
geom_bar(color="black") +
xlab("") +
theme(legend.position = "None")
p2 <-
results %>%
ggplot(aes(x=extract, y=lod, fill=factor(extract))) +
geom_boxplot() +
xlab("") +
theme(legend.position = "None")
p3 <-
results %>%
ggplot(aes(x=commod, y=lod, fill=factor(extract))) +
geom_boxplot() +
labs(fill="Extraction\nmethod") +
xlab("Commodity")
# Combine figures with library(cowplot)
cowplot::ggdraw() +
cowplot::draw_plot(p1, 0, .5, .5, .5) +
cowplot::draw_plot(p2, .5, .5, .5, .5) +
cowplot::draw_plot(p3, 0, 0, 1, .5) +
cowplot::draw_plot_label(c("A", "B", "C"), c(0, 0.5, 0), c(1, 1, 0.5), size = 15)
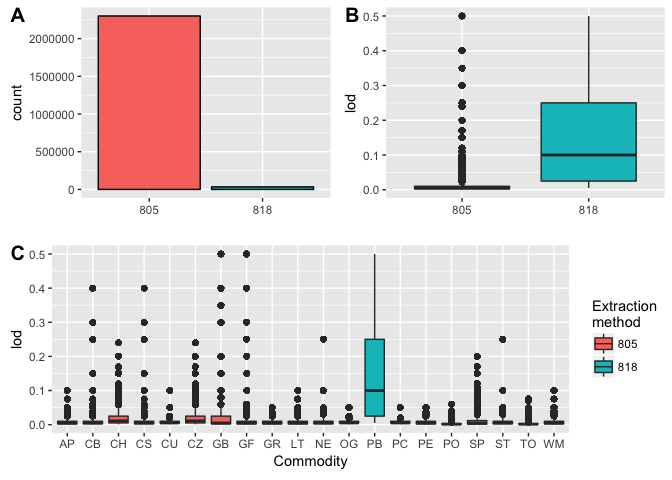
From the figure, the following insights could be drawn:
- [A] More samples are extracted by MDA Modified QuEChERS Method (805) than NSL Animal Tissue Extraction Method (818)
- [B] The limit of detection by MDA Modified QuEChERS Method (805) is lower, implying greater resolutions
- [C] “PB” is the only commodity extracted by NSL Animal Tissue Extraction Method (818)
Following the trail
PB had an unusually high Limit of Detection (LOD), I want to look into this further. When we compare the number of distinct levels per variable, we find only 1 lab does the NSL Animal Tissue Extraction Method (818).
## sample_pk commod commtype lab pestcode testclass
## 315 1 1 1 107 10
## concen lod conunit confmethod confmethod2 annotate
## 4 13 1 2 1 1
## quantitate mean extract determin
## 1 3 1 3
We next took a look at the LOD by Pesticide for each pesticide class type:
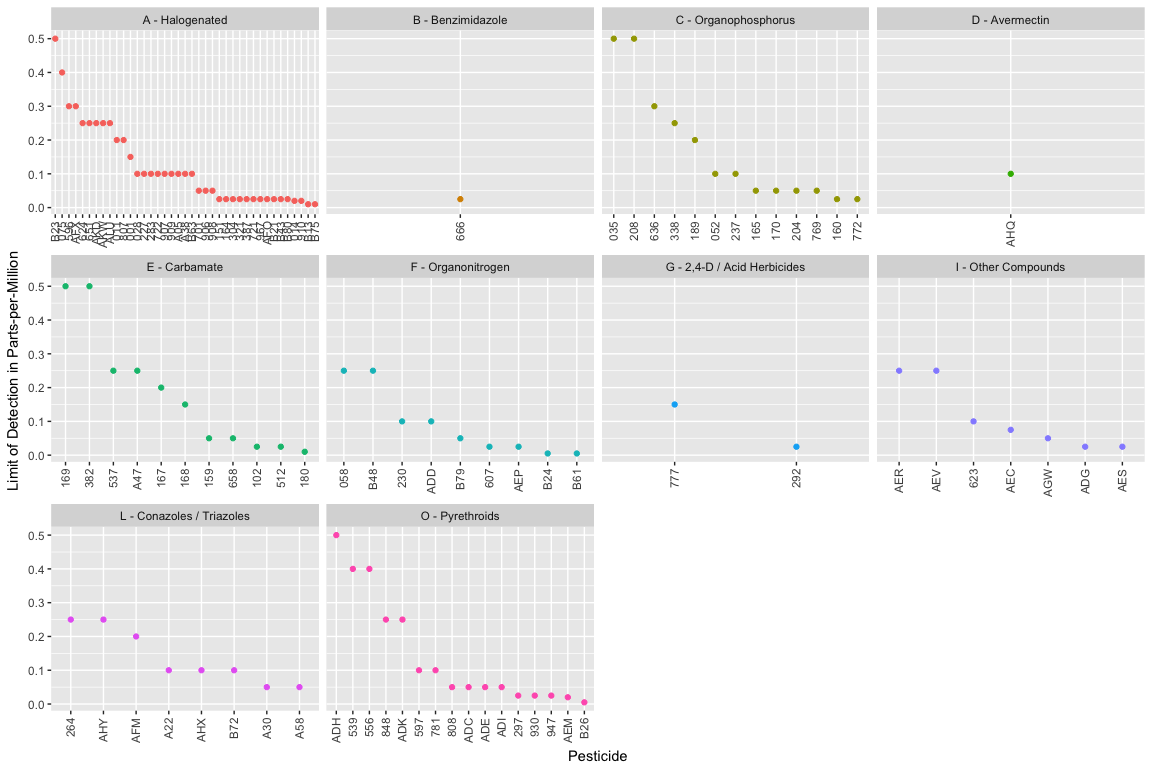
From this figure, we see (1) there are relatively a lot of Halogenated compounds and only a handful Benzimidazole, Avermectin, and 2,4-D / Acid Herbicides compounds; and (2) the pestcode, testclass, and LOD information are all linked.
(Initially I made a boxplot, but no boxes were drawn implying no variability.)
Reaching the right information
Since understanding the data more, I come to realize using only the complete cases is not the ideal way to go; instead, the columns with missing information (containing the pesticides concentrations) must be kept.
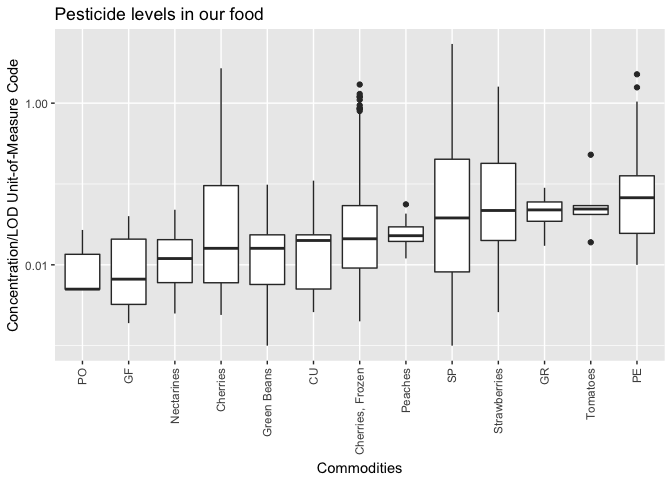
concen- Concentration/LOD Unit-of-Measure Codequantitate- QUANTITATION METHOD in 2015 PDP Analytical Resultsannotate-ANNOTATED INFORMATION in 2015 PDP Analytical Resultsconfmethod- CONFIRMATION METHOD in 2015 PDP Analytical Resultsconfmethod2
And using the the correct variable (ie. “concen”), we refine/corrected the figure from yesterday.