Yesterday I submitted a poster abstract to the ISMB 2016 conference in Orlando, Florida. This submission – submission #449 to category “Epigenetics”” – was a late submission, which meant that the results of submissions from rounds 1 are done and accepted abstracts have been released/listed onto the conference’s website.
I was curious.
My submission number is 449, this meant there were 448 submissions before mine. I want to know the following:
- How many abstracts have been accepted during round 1?
- Of the accepted abstracts in round 1, what proportion of them were categorized as “Epigenetic”?
16 possible categories
According to the ISMB website, there are 16 possible categories to which your abstract may belong.
| Item | Category/Link to abstracts | ID |
|---|---|---|
| 1. | Bioinformatics of Disease and Treatment | A |
| 2. | Comparative Genomics | B |
| 3. | Education | C |
| 4. | Epigenetics | D |
| 5. | Functional Genomics | E |
| 6. | Genome Organization and Annotation | F |
| 7. | Genetic Variation Analysis | G |
| 8. | Metagenomics | H |
| 9. | Open Science and Citizen Science | I |
| 10. | Pathogen informatics | J |
| 11. | Population Genetics Variation and Evolution | K |
| 12. | Protein Structure and Function Prediction and Analysis | L |
| 13. | Proteomics | M |
| 14. | Sequence Analysis | N |
| 15. | Systems Biology and Networks | O |
| 16. | Other | P |
And if we assume that people submit equally to all categories, we expect each category to contain 6.25% (100%/16) of the total number of abstracts.
Majority of posters are classified as “Sequence Analysis”
When we look at the number of abstracts to each category, we find the majority of them belong to “Sequence Analysis”.
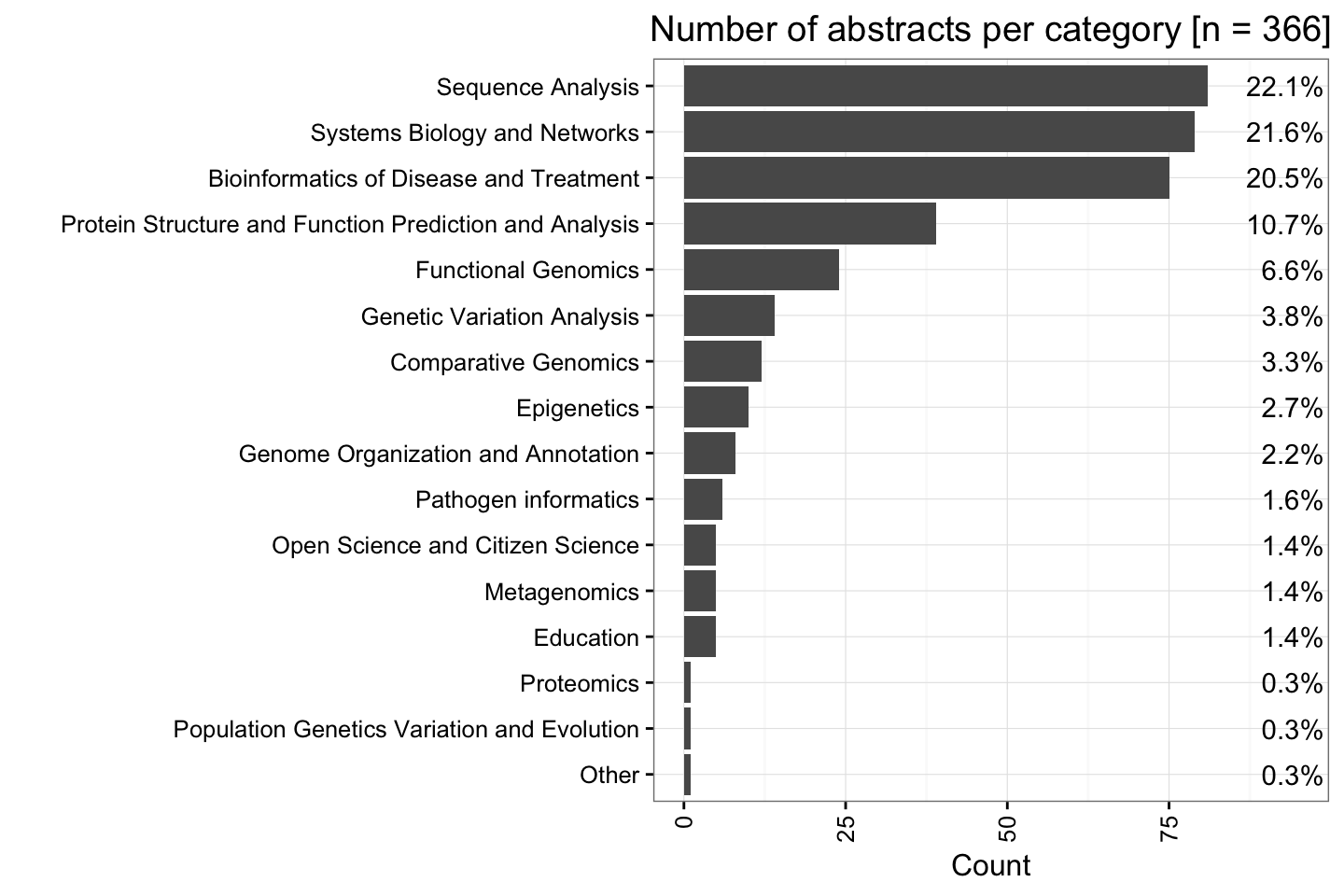
Here we also see – as of Fri, May 6, 2016 – there is a total of 366 abstracts accepted. From this total, only ~2.7% or 10 abstracts have been listed as “Epigenetics”.
Not bad news.
p.s.
I have also made available the list of 366 abstracts and the category to which they belong.